JupyterHub on WATGPU
JupyterHub allows you to easily run Jupyter notebooks on the cluster, providing you with another way to run interactive jobs to debug and test your code on the cluster. There is also the added benefit of being able to monitor your code's GPU utilization through the jupyterlab-nvdashboard extension that is enabled for all users, allowing you to understand if your code is efficiently using its allocated GPU.
Users who are looking to use Jupyter notebooks on WATGPU can securely sign in to WATGPU's JupyterHub at watgpu.cs.uwaterloo.ca/jupyterhub/ using their UWaterloo credentials and authenticate using Duo two-factor authentication.
After signing in, users can request resources by filling out a form that specifies, the compute node, number of CPU cores, memory (in GB), number of GPUs, and time.
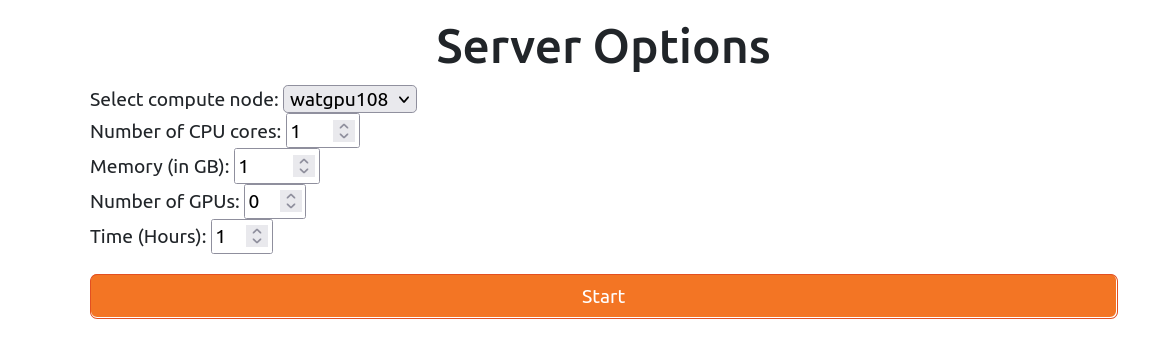
For now, Jupyter notebooks run through JupyterHub are submitted to the ALL partition, and have some limits on the session's resources:
- up to 16 CPU cores
- up to 128 GB of RAM
- up to 4 GPUs
- up to 4 hours of runtime
Users requesting Jupyter notebooks through JupyterHub with more than 8 CPU cores are recommended to submit these jobs to the watgpu408 compute node since that node has more CPU cores than other compute nodes.
Jupyter notebooks through JupyterHub are meant for debugging and testing your code, not for full production runs - we recommend submitting an sbatch job for full runs.
Virtual environments vs Jupyter kernels for Jupyter notebooks
Virtual environments, as explained in our section about virtual environments on the WATGPU cluster, are used as isolated directories that hold any associated libraries and packages required for a project. Jupyter kernels are used by Jupyter to run Jupyter notebooks in different virtual environments. You can use different virtual environments as different Jupyter kernels, so each Jupyter notebook can run with the libraries and packages specific to that environment.
When you create a Python virtual environment, it typically comes with its own Python interpreter that can be used to install different Python packages within the environment. To use that environment as a Jupyter kernel, you need to make it available to Jupyter by installing the ipykernel package and linking the environment to Jupyter, allowing you to select that environment as the kernel when you launch a Jupyter notebook.
Installing ipykernel and linking conda venvs to Jupyter
Activate the conda virtual environment you want to add as a kernel, then install ipykernel using conda:
(base) $ conda activate <conda-venv>
<conda-venv> $ conda install -c anaconda ipykernel
then add the conda virtual environment as a kernel:
<conda-venv> $ python -m ipykernel install --user --name=<conda-venv>
Installing ipykernel and linking pip venvs to Jupyter
Activate the pip virtual environment you want to add as a kernel, the install ipykernel using pip:
$ source <venv-name>/bin/activate
<venv-name> $ pip install ipykernel
then add the pip virtual environment as a kernel:
<venv-name> $ python -m ipykernel install --user --name=<venv-name>
You can then launch a Jupyter notebook from the directory that contains the pip virtual environment directory.